Proteomics Tools
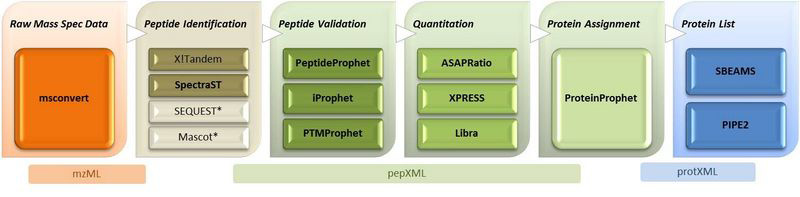
The Trans-Proteomic Pipeline (TPP) includes all of the steps of the ISB MS/MS analysis pipeline, after the database search. Most users will only need to download the TPP distrubtion.
Download the TPP Installer for Windows
The individual tools that make up the TPP, as well as a few which are not included in the TPP distrubution, are categorized below.
Please see the SPC Tools Wiki for more information, including download information.
- Input Processing / Data Handling
- mzXML – standard format for mass-spectrometry data
- ReAdW – Thermo Xcalibur-to-mzXML converter
- massWolf – Waters MassLynx raw-to-mzXML converter
- mzWiff – ABI/MDS Sciex Analyst raw-to-mzXML converter
- mzBruker – Bruker raw-to-mzXML converter
- RAP, JRAP, RAMP – mzXML data parsers
- validateXML – mzXML validation script
- MzXML2Search – mzXML to SEQUEST dta, MASCOT generic and Micromass pkl converter
- mzXMLViewer – viewer for displaying mzXML data
- InsilicosViewer – viewer for displaying mzXML data
- readmzXML – mzXML parser based on RAMP
- Database and Spectral Library Search
- SpectraSTTM – spectral library search engine provided by TPP
- Comet – open-source database search engine provided with TPP
- X!Tandem – open-source database search engine provided with TPP/li>
- Probability Assignment and Validation
- PeptideProphetTM – validation of peptide identifications made by tandem mass spectrometry (MS/MS) and database searching; probabilities are assigned to the peptide identifications made by programs like SEQUEST or MASCOT
- iProphetTM – validation of distinct peptide sequences; can also combine search results of multiple search engines
- ProteinProphetTM – statistical model for validation of peptide identifications at the protein level
- Protein Quantification
- Protein ID Curation
- QualScore— analyze unassigned but high quality spectra.
- PepXMLViewer – peptide dataset interaction
- Pep3D – viewer for LC-MS and LC-MS/MS results
- Out2Summary – converter of SEQUEST and TurboSEQUEST *.out files for TPP processing into a single HTML-SUMMARY file ready for use with INTERACT
- Protein Crosslinking
- XLink – XLink suite
- Miscellaneous
- PeptideSieve – A tool for predicting proteotypic peptides
- SubsetDB – FASTA manipulation
- LC-MS Analysis
- Interpretation